Contents
- Image compression by Reduce Basis Decomposition
- Face recognition by Reduced Basis Decomposition.
- Formation of training set and test set
- Testing the eigen faces and calculate the success rate
- Plotting the error rates
Called Functions
Image compression by Reduce Basis Decomposition
Author: Yanlai Chen
Version: 1.0
Date: 03/20/2015
thispic = imread('Figures/lena.png'); compratio = 10; figshrink = 1; m = size(thispic,1); n = size(thispic,2); num = floor(min(m,n)/compratio); fprintf('Shrinking from (%d, %d) to (%d, %d) and (%d, %d)\n',m,n,m,num,num,n); datad3 = size(thispic,3);
Shrinking from (512, 512) to (512, 51) and (51, 512)
Displaying the original picture as is
figure('Position',[1,1,1+n/figshrink,1+m/figshrink]) if(datad3 == 1) imagesc(thispic);colormap(gray);drawnow; else image(thispic); title('Original image');drawnow; end axis off

Handling it by RBD on each component
bases = zeros(m,num,datad3); TransMat = zeros(num,n,datad3); tic for i=1:datad3 data = double(thispic(:,:,i)); [bases(:,:,i), TransMat(:,:,i)] = RBD(data, 1e-20,num); end t_rb = toc;
Recovering the compressed image from its RBD bases and displaying it
B10 = zeros(m,n,datad3); for j=1:datad3 B10(:,:,j) = bases(:,:,j)*TransMat(:,:,j); end ScaleAndSave(B10,m,n,datad3,'By RB component-wise compression',figshrink);

Handling it by SVDS
U = zeros(m,num,datad3); S = zeros(num,num,datad3); V = zeros(n,num,datad3); U = zeros(m,num,datad3); S = zeros(num,num,datad3); V = zeros(n,num,datad3); tic for i=1:datad3 data = double(thispic(:,:,i)); [U(:,:,i), S(:,:,i), V(:,:,i)] = svds(data,num); end t_svds = toc;
Handling it by SVD
U = zeros(m,m,datad3); S = zeros(m,n,datad3); V = zeros(n,n,datad3); tic for i=1:datad3 data = double(thispic(:,:,i)); [U(:,:,i), S(:,:,i), V(:,:,i)] = svd(data); end t_svd = toc;
Recovering the compressed image from its SVDD bases and displaying it
A10 = zeros(m,n,datad3); for j=1:datad3 for i=1:num A10(:,:,j) = A10(:,:,j) + S(i,i,j)*U(:,i,j)*V(:,i,j)'; end end ScaleAndSave(A10,m,n,datad3,'By SVD',figshrink);

RBD image compression is faster than SVD
fprintf('Time spend on (svd, svds, rbd) relative to svd is (%e, %e, %e)\n',t_svd/t_svd, t_svds/t_svd, t_rb/t_svd);
Time spend on (svd, svds, rbd) relative to svd is (1.000000e+00, 3.907092e+00, 2.636303e-01)
Face recognition by Reduced Basis Decomposition.
We test our algorithm on the UMIST data set available at Roweis' website http://www.cs.nyu.edu/ roweis/data.html
load('Figures/umist_cropped.mat');
totalp = 20;
It contains 575 cropped facial images (20 people under different poses) Here is a snapshot of the database
umist_snap = imread('Figures/umist_snapshot.png'); figure('Position',[1,1,1+size(umist_snap,2),1+size(umist_snap,1)]) imagesc(umist_snap);colormap(gray);drawnow axis off

Preliminary setup
trainnum: number of views randomly take per subject for training.
totalrun: number of rounds of formations of training and test sets
basnum: number of total eigen faces taken from the whole training set
avgrate: the average success rate when totalrun rounds are done
trainnum = 10; totalrun = 2; avgrate = []; for basnum = 5:10:100 thisrate = 0; for runnum = 1: totalrun
trainpic2d = [];
tp_ind = zeros(totalp,trainnum);
testpic2d = [];
Formation of training set and test set
We first lineup all the pictures and then randomly form the training set and test set.
for i=1:totalp pic_pi = facedat{i}; num_pi = size(pic_pi,3); [nr,nc] = size(pic_pi(:,:,1)); pic2d = zeros(nr*nc,num_pi); for j=1:num_pi pic2d(:,j) = reshape(pic_pi(:,:,j),nr*nc,1); end pic2ds{i} = pic2d; tmp = 0; while (tmp < trainnum) tn = randi(num_pi); if(size(find(tp_ind(i,1:tmp) == tn),2) == 0) tp_ind(i,tmp+1) = tn; tmp = tmp+1; trainpic2d = [trainpic2d double(reshape(pic_pi(:,:,tn),nr*nc,1))]; end end end
Method number 1: A simple PCA approach to find the eigen faces
tic
PCA_FR(trainpic2d);
t_pca = toc;
Method number 2: RBD to find the (basnum) eigen faces
tic
totalmean = mean(trainpic2d,2);
trainpic2d = trainpic2d - totalmean*ones(1,size(trainpic2d,2));
[bases, TransMat] = RBD(trainpic2d, 1e-20,basnum,1,zeros(nr*nc,1));
t_rbd = toc;
The comparison in efficiency
fprintf('PCA time over RBD time when %d eigen faces are used is %f\n',basnum,t_pca/t_rbd);
PCA time over RBD time when 5 eigen faces are used is 2.608177
PCA time over RBD time when 15 eigen faces are used is 1.893146
PCA time over RBD time when 25 eigen faces are used is 1.483418
PCA time over RBD time when 35 eigen faces are used is 1.071684
PCA time over RBD time when 45 eigen faces are used is 0.845395
PCA time over RBD time when 55 eigen faces are used is 0.702518
PCA time over RBD time when 65 eigen faces are used is 0.559466
PCA time over RBD time when 75 eigen faces are used is 0.450852
PCA time over RBD time when 85 eigen faces are used is 0.372011
PCA time over RBD time when 95 eigen faces are used is 0.321043
Testing the eigen faces and calculate the success rate
totaltest: the total number of tests you want to perform
matched = 0;
totaltest = 100;
for i=1:totaltest
pnum = randi(totalp);
% picnum = tp_ind(pnum,randi(trainnum)); % test those in the
% training set, success rate should be 100%
% test those that are not in the training set
intraining = 1;
while(intraining == 1)
picnum = randi(size(pic2ds{pnum},2));
if(size(find(tp_ind(pnum,:) == picnum),2) == 0)
intraining = 0;
end
end
thispic = pic2ds{pnum}(:,picnum) - totalmean;
all_err = 1e14;
bestall = 0;
coe = bases'*thispic;
for j=1:size(TransMat,2)
jth_err = norm(coe - TransMat(:,j));
if(jth_err <all_err)
all_err = jth_err;
bestall = j;
end
end
bestall = ceil(bestall/(trainnum+1e-10));
if(bestall == pnum)
matched = matched+1;
end
end
thisrate = thisrate + matched/totaltest;
end thisrate = thisrate/totalrun; fprintf('The average success rate using %d eigen faces for %d runs of recognizing %d images is %f\n',basnum,totalrun,totaltest,thisrate) avgrate = [avgrate thisrate]; end
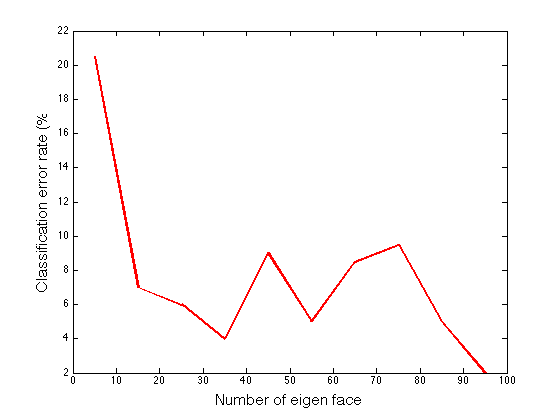
The average success rate using 5 eigen faces for 2 runs of recognizing 100 images is 0.795000 The average success rate using 15 eigen faces for 2 runs of recognizing 100 images is 0.930000 The average success rate using 25 eigen faces for 2 runs of recognizing 100 images is 0.940000 The average success rate using 35 eigen faces for 2 runs of recognizing 100 images is 0.960000 The average success rate using 45 eigen faces for 2 runs of recognizing 100 images is 0.910000 The average success rate using 55 eigen faces for 2 runs of recognizing 100 images is 0.950000 The average success rate using 65 eigen faces for 2 runs of recognizing 100 images is 0.915000 The average success rate using 75 eigen faces for 2 runs of recognizing 100 images is 0.905000 The average success rate using 85 eigen faces for 2 runs of recognizing 100 images is 0.950000 The average success rate using 95 eigen faces for 2 runs of recognizing 100 images is 0.980000
Plotting the error rates
figure() plot(5:10:100,100 - avgrate*100,'-r','LineWidth',2,'MarkerEdgeColor','k','MarkerFaceColor','g','MarkerSize',10); xlabel('Number of eigen faces','fontsize',15); ylabel('Classification error rate (%)','fontsize',15);